Project Summary
RNA expression profiling
- RNA expression profiling
- Developing earshoots
- Leaf and seed tissues during grain-fill
Genetic mapping of phenotypes associated with NUE
- Agronomic traits (biomass, N accumulation and partitioning)
- Metabolites (nitrate, amino acids)
- Gene expression (eQTL)/enzyme activities (aQTL)
Candidate gene validation
- Coincidence among QTL and differentially expressed genes.
- Functional testing of genes
Developing Country Collaboration with the International Institute for Tropical Agriculture (IITA)
RNA Expression Profiling to Discover N-Responsive Genes
Our initial approach to identifying "Nitrogenes" is to compare gene expression and physiological traits of maize germplasm grown with different levels of N fertility. Maize NUE is best studied in hybrids (the cross of two inbred parents), because N responses are greater in hybrids and hybrids are what farmers grow. Our comparisons focus on hybrids that have the inbred line B73 as one parent, because the B73 line is the focus of the maize genome sequencing effort. We are using four distinct sets of germplasm in these comparisons:
- B73 x Mo17 hybrid, which is representative of hybrids grown in the U.S. Corn Belt
- Diversity Hybrids, where a set of inbreds selected to represent the range of genetic diversity in maize are crossed to B73.
- IHP Hybrids, inbreds derived from Illinois High Protein (IHP) crossed to B73. IHP is a strain subjected to more than a century of genetic selection for grain protein concentration, which also has dramatically altered N metabolism in the plant.
- Hybrids from an ongoing breeding program to improve NUE at the International Institute for Tropical Agriculture. These hybrids are adapted to the low fertility soils of West and Central Africa.
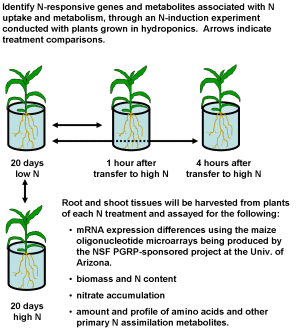
To compare gene expression and physiological responses to N, we grow maize with different levels of N
in both greenhouse hydroponics and field experiments. These environments maximize the probability of
discovering genes associated with NUE.
|
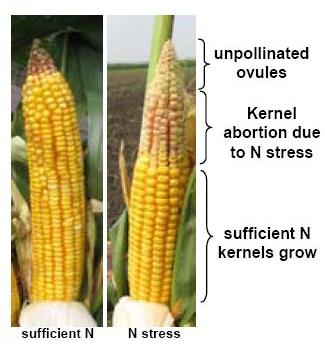
The primary physiological response to N stress that is most highly correlated with grain yield and NUE is a reduction in the number of fully developed kernels, which is most pronounced at the tip of the ear. The tip portion (25%) of developing earshoots was sampled from B73 x Mo17 hybrid plants grown in the field either with or without supplemental N. mRNA isolated from these tissues was hybridized to microarrays produced by the Maize Oligonucleotide Microarray Project at the University of Arizona. ~ 9,000 features passed quality check for both N treatments, where signal intensities for both dyes was above background on all eight replicate slides. 326 features show statistically significant expression differences in response to N
|
||||||||||||||||||||||||
|
|||||||||||||||||||||||||
|
|||||||||||||||||||||||||
Identifying QTL Controlling NUE Traits
Another approach to identify genes that control NUE in maize is to associate small segments of chromosomes to measurable traits that contribute to NUE. Such chromosomal segments are named quantitative trait loci, or QTL. The general strategy to find QTL is to produce a population of lines that have their chromosomes shuffled through genetic recombination, where alleles are exchanged among parents without changing gene order along the chromosome. Molecular markers are then used to define the exchanged segments and construct a genetic map. Traits are measured in individuals from the population, and statistical techniques are used to identify those chromosomal regions that are associated with variation for the trait. Once defined, the genes within the QTL segments that cause the trait difference can be identified.
The intermated B73 x Mo17 recombinant inbred lines (IBMRIs) are the highest resolution mapping population available in maize. The Illinois High Protein1 (IHP1) inbred line exhibits the greatest capacity for N uptake measured in maize. Hybrids between 250 of the IBMRIs crossed to IHP1 were grown in 2006 and 2007, with each hybrid receiving either no or 250 kg/ha of supplemental N in adjacent plots within a replicated field design Searches for QTL are being conducted for the following traits:
- NUE, N uptake, N utilization by developing grain
- stover and grain biomass at anthesis, harvest
- stover and grain N concentration at anthesis, harvest.
- N metabolites (nitrate, amino acids) in leaves and earshoots
- grain yield and its components, kernel number and weight
- RNA expression for key N metabolism genes (eQTL)
- Activities of enzymes for N assimilation and C/N balance.
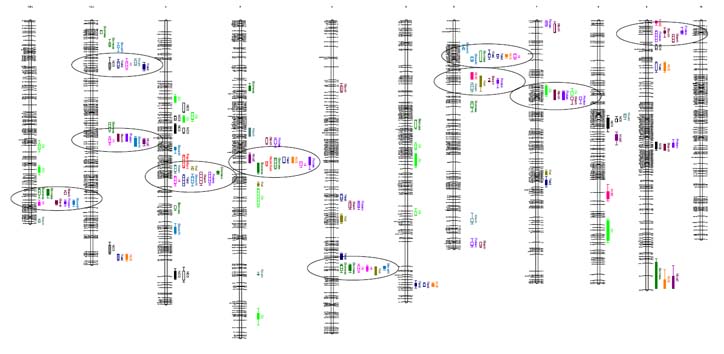
Maize IBM genetic map showing locations of QTL for N-responsive traits. Circled chromosomal regions harbor QTL for multiple traits. Detailed maps to follow in journal articles that are in preparation.
Validation of Candidate Genes
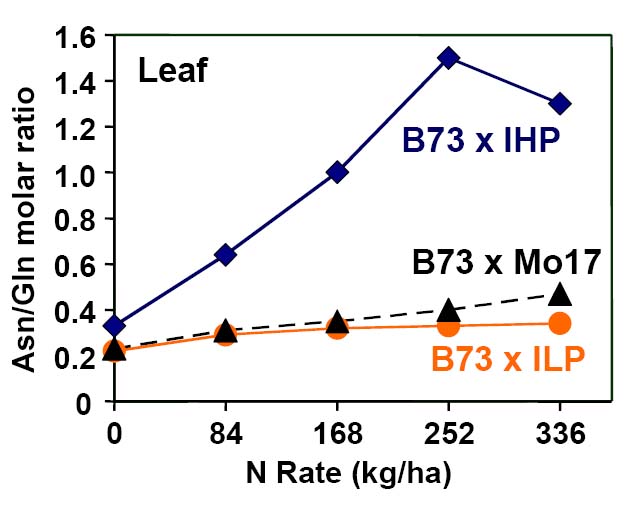
Leaf amino acid profile changes in response to N among B73 x IHP1, B73 x ILP1, and B73 x Mo17 hybrids. The transport and storage amino acid asparagine (Asn) increases relative to metabolically active glutamine (Gln) in response to N and the IHP genotype.
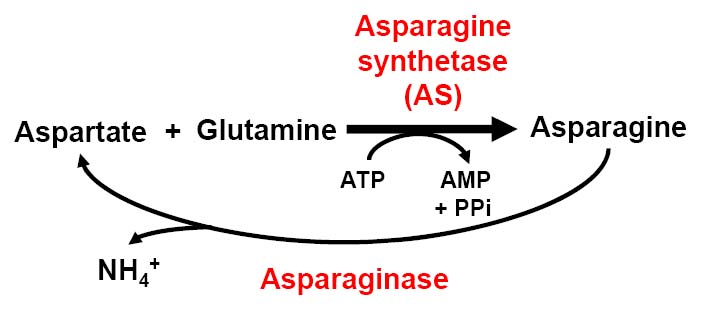
The metabolic pahtway for asparagine accumulation.

B73 and IHP1 plants were grown in hydroponics under either Ndeficient (1 mM NO3 -) or Nsufficient (14 mM NO3 -) conditions. Leaves were sampled from these plants during both the day and night. RNA was isolated and the relative expression of genes encoding an asparagine synthetase (AS2) or asparaginase isoform was assayed by qRT-PCR.
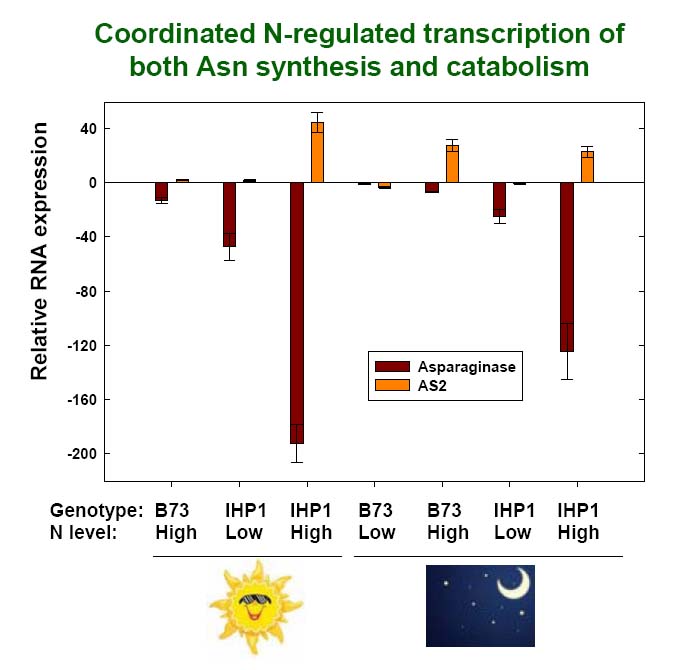
Quantitative Real-Time PCR
analysis of NitroGenes:
AS2 RNA expression
increases in response to N
and is also elevated in IHP1
relative to B73, particularly
during the day. Conversely,
asparaginase RNA
expression is repressed by
N, and is greatly reduced in
IHP1 compared to B73.
Developing Country Collaboration with the International Institute for Tropical Agriculture (IITA)
Maize is a major cereal crop in West and Central Africa, accounting for more than 20% of domestic food production. Not only has maize replaced sorghum and millet as food staples, but it is fast becoming an important source of cash for smallholder farmers (Smith et al., 1997). The low N content in soils of the Western and Central African savannas, coupled with the high costs of N fertilizer, often make N the limiting nutrient for maize production. As a result, N-deficiency is common in the maize crop produced in this geographical region.
The International Institute for Tropical Agriculture (IITA), based in Nigeria, is a member of the CGIAR crop improvement centers. In an ongoing effort to improve the NUE of maize varieties adapted to West and Central Africa, IITA has operated a breeding program focused on NUE for nearly 15 years, coordinated most recently by Dr. Abebe Menkir. This breeding program is the main source of germplasm for improved varieties in Western and Central Africa. However, IITA and other institutions in Africa have limited facilities and expertise to take advantage of resources for maize genomics research. We are develop avenues for information exchange between our project and scientists at IITA that will allow us to determine whether candidate genes identified through the analysis of B73 x Mo17 and IBMRI x IHP1 hybrids are relevant to improving NUE in maize germplasm adapted to West and Central Africa.
Our collaboration has four general objectives:
- to support reciprocal visits between a graduate student working with IITA collaborators and the project scientists, where the graduate student will be trained in sampling and analysis methods for NUE and its component traits
- to assess in field trials the physiological strategies being employed by IITA maize varieties with improved NUE, including amino acid profiles under low N conditions
- to evaluate IHP1 as a germplasm source for improving NUE in African maize
- to conduct RNA expression profiling to assess patterns of N-responsive gene expression and genetic variability among the best IITA maize varieties for NUE.
This collaboration will provide a number of significant benefits to agricultural production in Western and Central Africa. The exchange of ideas, approaches, and data will further enhance the breeding efforts of IITA to increase maize NUE. Because IITA is a major provider of maize germplasm to sub- Saharan Africa, any improved varieties will be rapidly disseminated to farmers in these areas. The adoption of maize varieties with better NUE will raise yields of the maize crop in West and Central Africa while using less supplemental N–a prospect that not only reduces input and energy costs, but also decreases the potential N pollution of scarce water supplies in the region.


