Identifying QTL Controlling NUE Traits
Another approach to identify genes that control NUE in maize is to associate small segments of chromosomes to measurable traits that contribute to NUE. Such chromosomal segments are named quantitative trait loci, or QTL. The general strategy to find QTL is to produce a population of lines that have their chromosomes shuffled through genetic recombination, where alleles are exchanged among parents without changing gene order along the chromosome. Molecular markers are then used to define the exchanged segments and construct a genetic map. Traits are measured in individuals from the population, and statistical techniques are used to identify those chromosomal regions that are associated with variation for the trait. Once defined, the genes within the QTL segments that cause the trait difference can be identified.
The intermated B73 x Mo17 recombinant inbred lines (IBMRIs) are the highest resolution mapping population available in maize. The Illinois High Protein1 (IHP1) inbred line exhibits the greatest capacity for N uptake measured in maize. Hybrids between 250 of the IBMRIs crossed to IHP1 were grown in 2006 and 2007, with each hybrid receiving either no or 250 kg/ha of supplemental N in adjacent plots within a replicated field design Searches for QTL are being conducted for the following traits:
- NUE, N uptake, N utilization by developing grain
- stover and grain biomass at anthesis, harvest
- stover and grain N concentration at anthesis, harvest.
- N metabolites (nitrate, amino acids) in leaves and earshoots
- grain yield and its components, kernel number and weight
- RNA expression for key N metabolism genes (eQTL)
- Activities of enzymes for N assimilation and C/N balance.
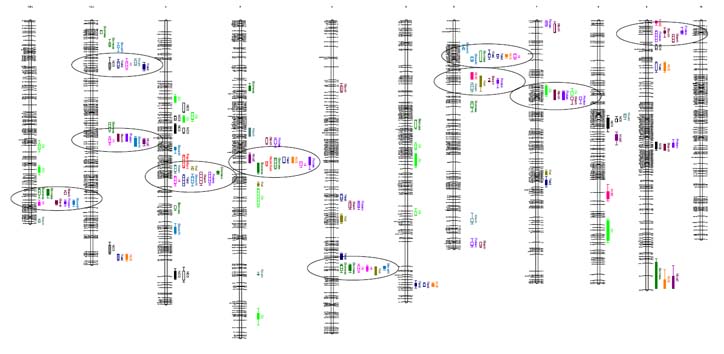
Maize IBM genetic map showing locations of QTL for N-responsive traits. Circled chromosomal regions harbor QTL for multiple traits. Detailed maps to follow in journal articles that are in preparation.


